Genos
Collection
2 items
•
Updated
•
1
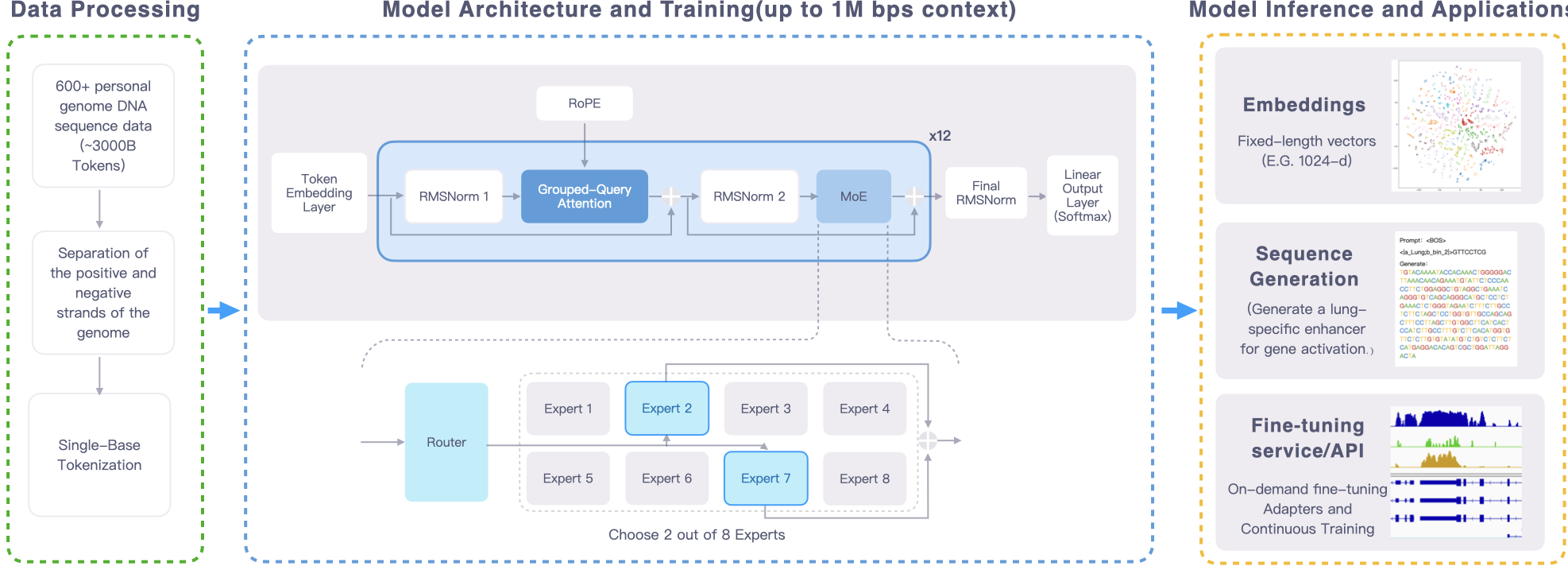
Genos, as a foundational model in the field of human genomics, trained on hundreds of high-quality genome reference data, has achieved the ability to contextually model human genome sequences up to millions of base pairs. Through single-base resolution learning, this model possesses the capability to identify hidden deep sequence patterns and functional features within genomes, providing scientists with a new research method that connects genetic information with life activities.
For instructions, details, and examples, please refer to the Genos GitHub.
Below are the data volume of our model training and related parameters.
| Model Specification | Genos 1.2B | Genos 10B |
|---|---|---|
| Model Scale | ||
| Total Parameters | 1.2B | 10B |
| Activated Parameters | 0.33B | 2.87B |
| Trained Tokens | 1600 B | 2200 B |
| Architecture | ||
| Architecture Type | MoE | MoE |
| Number of Experts | 8 | 8 |
| Selected Experts per Token | 2 | 2 |
| Number of Layers | 12 | 12 |
| Attention Hidden Dimension | 1024 | 4096 |
| Number of Attention Heads | 16 | 16 |
| MoE Hidden Dimension (per Expert) | 4096 | 8192 |
| Vocabulary Size | 128 (padded) | 256 (padded) |
| Context Length | up to 1M | up to 1M |
Genos 1.2B and 10B checkpoints are available here:
We also provide checkpoints trained under the Megatron-LM framework: